Journal of Traditional Chinese Medicine ›› 2025, Vol. 45 ›› Issue (6): 1342-1352.DOI: 10.19852/j.cnki.jtcm.2025.06.013
• Original Articles • Previous Articles Next Articles
Network pharmacology combined with ultra-high-performance liquid chromatography-quadrupole time-of-flight mass spectrometry method to explore the mechanism of Shizhi Fang (矢志方) in treating uric acid nephropathy mice
YANG Feng1,2,3,4, ZHANG Xuming1,2,3,4, WU Zhiyuan1,2,3,4, GAO Jiandong1,2,3,4( )
)
- 1 Department of Nephrology, Shuguang Hospital Affiliated to Shanghai University of Traditional Chinese Medicine, Shanghai 200120, China
2 TCM Institute of Kidney Disease, Shanghai University of Traditional Chinese Medicine, Shanghai 200120, China
3 Key Laboratory of Liver and Kidney Diseases, Ministry of Education, Shanghai University of Traditional Chinese Medicine, Shanghai 200120, China
4 Shanghai Key Laboratory of Traditional Chinese Clinical Medicine, Shanghai University of Traditional Chinese Medicine, Shanghai 200120, China
-
Received:2024-11-22Accepted:2025-03-10Online:2025-12-15Published:2025-11-24 -
Contact:Prof. GAO Jiandong, Department of Nephrology, Shuguang Hospital Affiliated to Shanghai University of Traditional Chinese Medicine, Shanghai 200120, China, jiandong. gao@shutcm.edu.cn, Telephone: +86-15572517091 -
Supported by:Mechanistic Study on How Shizhi Fang Promotes Mitocytosis of Renal Tubular Epithelial Cells to Alleviate Hyperuricemia-Induced Kidney Injury(82274415);Mechanistic Study on Shizhi Fang in Lowering Uric Acid Based on Extracellular Signal-regulated Kinases 1 and 2-mediated Transcriptional Regulation of the Uric Acid Transporter Urate Transporter 1(82474434)
Cite this article
YANG Feng, ZHANG Xuming, WU Zhiyuan, GAO Jiandong. Network pharmacology combined with ultra-high-performance liquid chromatography-quadrupole time-of-flight mass spectrometry method to explore the mechanism of Shizhi Fang (矢志方) in treating uric acid nephropathy mice[J]. Journal of Traditional Chinese Medicine, 2025, 45(6): 1342-1352.
share this article
| Peak No. | Rt (min) | Selected-ion | Measured mass (m/z) | Ture values (m/z) | Error (ppm) | Formula | Molecu-lar weight | Name | MS/MS | Classification |
|---|---|---|---|---|---|---|---|---|---|---|
| P1 | 2.37 | [M-H]- | 424.0380 | 424.0378 | 0.6 | C14H19NO10S2 | 425.05 | Sinalbin | 259.0107; 236.0911; 96.9607 | Sinapis alba L. |
| P2 | 11.10 | [M-H]- | 373.1137 | 373.1140 | -0.9 | C16H22O10 | 374.12 | Geniposidic acid | 211.0638; 123.0464 | Plantago indica L. |
| P3 | 12.53 | [M+H]+ | 224.1391 | 224.1394 | -1.1 | C11H17N3O2 | 223.13 | Plumbagine B or isomer | / | Plantago indica L. |
| P4 | 14.48 | [M+H]+ | 224.1384 | 224.1394 | -4.3 | C11H17N3O2 | 223.13 | Plumbagine B or isomer | / | Plantago indica L. |
| P5 | 17.58 | [M+H]+ | 226.1550 | 226.1550 | 0.0 | C11H19N3O2 | 225.15 | Plantagoguanidinic acid | 208.1456;84.0554 | Plantago indica L. |
| P6 | 20.07 | [M+H]+ | 314.2075 | 314.2074 | 0.2 | C15H27N3O4 | 313.20 | Plumbagine D | 296.1985; 172.1067; 84.0555 | Plantago indica L. |
| P7 | 23.78 | M+ | 310.1643 | 310.1649 | -1.9 | C16H24NO5+ | 310.17 | Sinapine | / | Sinapis alba L. |
| P8 | 35.94 | [M-H]- | 223.0613 | 223.0612 | 0.5 | C11H12O5 | 224.07 | Sinapic acid | / | Sinapis alba L. |
| P9 | 41.31 | [M-H]- | 563.1413 | 563.1406 | 1.2 | C26H28O14 | 564.15 | Isovitexin 2"-O-arabinoside | / | Gypsophila vaccaria (L.) Sm. |
Table 1 Analysis and identification of prototype compounds in rat plasma of SZF based on UPLC-Q-TOF-MS
| Peak No. | Rt (min) | Selected-ion | Measured mass (m/z) | Ture values (m/z) | Error (ppm) | Formula | Molecu-lar weight | Name | MS/MS | Classification |
|---|---|---|---|---|---|---|---|---|---|---|
| P1 | 2.37 | [M-H]- | 424.0380 | 424.0378 | 0.6 | C14H19NO10S2 | 425.05 | Sinalbin | 259.0107; 236.0911; 96.9607 | Sinapis alba L. |
| P2 | 11.10 | [M-H]- | 373.1137 | 373.1140 | -0.9 | C16H22O10 | 374.12 | Geniposidic acid | 211.0638; 123.0464 | Plantago indica L. |
| P3 | 12.53 | [M+H]+ | 224.1391 | 224.1394 | -1.1 | C11H17N3O2 | 223.13 | Plumbagine B or isomer | / | Plantago indica L. |
| P4 | 14.48 | [M+H]+ | 224.1384 | 224.1394 | -4.3 | C11H17N3O2 | 223.13 | Plumbagine B or isomer | / | Plantago indica L. |
| P5 | 17.58 | [M+H]+ | 226.1550 | 226.1550 | 0.0 | C11H19N3O2 | 225.15 | Plantagoguanidinic acid | 208.1456;84.0554 | Plantago indica L. |
| P6 | 20.07 | [M+H]+ | 314.2075 | 314.2074 | 0.2 | C15H27N3O4 | 313.20 | Plumbagine D | 296.1985; 172.1067; 84.0555 | Plantago indica L. |
| P7 | 23.78 | M+ | 310.1643 | 310.1649 | -1.9 | C16H24NO5+ | 310.17 | Sinapine | / | Sinapis alba L. |
| P8 | 35.94 | [M-H]- | 223.0613 | 223.0612 | 0.5 | C11H12O5 | 224.07 | Sinapic acid | / | Sinapis alba L. |
| P9 | 41.31 | [M-H]- | 563.1413 | 563.1406 | 1.2 | C26H28O14 | 564.15 | Isovitexin 2"-O-arabinoside | / | Gypsophila vaccaria (L.) Sm. |
| Peak No. | Rt (min) | Selected- ion | Measured mass (m/z) | Ture values (m/z) | Error (ppm) | Formula | Molecular weight | Name | MS/MS |
|---|---|---|---|---|---|---|---|---|---|
| M1 | 4.14 | [M-H]- | 233.0120 | 233.0125 | 0.3 | C8H10O6S | 234.02 | Hydroxytyrosol+sulfation | 233.0120;153.0555;123.0449 |
| M2 | 4.54 | [M-H]- | 233.0125 | 233.0125 | 0.0 | C8H10O6S | 234.02 | Hydroxytyrosol+sulfation | 233.0109;153.0551;123.0447 |
| M3 | 5.70 | [M-H]- | 261.0081 | 261.0074 | 2.5 | C9H10O7S | 262.01 | Caffeic acid+hydrogenation+sulfation | / |
| M4 | 7.59 | [M-H]- | 247.0281 | 247.0282 | -0.3 | C9H12O6S | 248.04 | Hydroxytyrosol+methylation+sulfation | / |
| M5 | 8.44 | [M-H]- | 357.0828 | 357.0827 | 0.2 | C15H18O10 | 358.09 | Homovanillic acid+glucuronidation | 181.0494;175.0262;137.0605;113.0230 |
| M6 | 9.95 | [M-H]- | 245.0124 | 245.0125 | -0.5 | C9H10O6S | 246.02 | 4-Hydroxybenzoylcholine-(C3H8N+)+sulfation | 245.011;6165.0554;121.0651 |
| M7 | 11.51 | [M-H]- | 343.1034 | 343.1035 | -0.1 | C15H20O9 | 344.11 | Homovanillic alcohol+glucuronidation | 343.1300;167.0690;152.0490;113.0240 |
| M8 | 12.50 | [M-H]- | 275.0230 | 275.0231 | -0.4 | C10H12O7S | 276.03 | Genipinic acid+dehydroxylation+sulfation | / |
| M9 | 12.74 | [M-H]- | 371.0981 | 371.0984 | -0.7 | C16H20O10 | 372.11 | Genipinic acid+dehydroxylation+glucuronidation | 177.0519 |
| M10 | 13.62 | [M-H]- | 258.9917 | 258.9918 | -0.4 | C9H8O7S | 260.00 | Caffeic acid+sulfation | / |
| M11 | 13.73 | [M-H]- | 275.0229 | 275.0231 | -0.7 | C10H12O7S | 276.03 | Dihydroferulic acid+sulfation | 275.0255;195.0689;136.0514 |
| M12 | 15.23 | [M-H]- | 327.0721 | 327.0722 | -0.2 | C14H16O9 | 328.08 | 4-Hydroxybenzoylcholine-(C4H10N+)+glucuronidation | 151.0396;136.0164;113.0242 |
| M13 | 16.14 | [M-H]- | 245.0124 | 245.0125 | -0.5 | C9H10O6S | 246.02 | 4-Hydroxybenzoylcholine-(C3H8N+)+sulfation | 165.0550;150.0330 |
| M14 | 17.29 | [M-H]- | 303.0179 | 303.0180 | -0.4 | C11H12O8S | 304.03 | Sinapic acid+sulfation | / |
| M15 | 17.29 | [M-H]- | 303.0182 | 303.0180 | 0.6 | C11H12O8S | 304.03 | Sinapic acid+sulfation | / |
| M16 | 18.23 | [M-H]- | 273.0065 | 273.0074 | -3.5 | C10H10O7S | 274.01 | Ferulic acid+sulfation | 193.0505;178.0263;134.0368 |
| M17 | 19.53 | [M-H]- | 399.0930 | 399.0933 | -0.7 | C17H20O11 | 400.10 | Sinapic acid+glucuronidation | / |
| M18 | 19.62 | [M-H]- | 273.0077 | 273.0074 | 0.9 | C10H10O7S | 274.01 | Caffeic acid+methylation+sulfation | / |
| M19 | 20.10 | [M-H]- | 273.0075 | 273.0074 | 0.2 | C10H10O7S | 274.01 | Ferulic acid+sulfation | 193.0508;178.0275;134.0382 |
| M20 | 20.69 | [M-H]- | 399.0967 | 399.0933 | 8.6 | C17H20O11 | 400.10 | Sinapic acid+glucuronidation | / |
| M21 | 29.35 | [M-H]- | 621.1120 | 621.1097 | 3.7 | C27H26O17 | 622.12 | Apigenin+diglucuronidation | / |
| M22 | 34.77 | [M-H]- | 539.0508 | 539.0501 | 1.3 | C22H20O14S | 540.06 | Apigenin+methylation+glucuronidation+sulfation | 363.0188;283.0599;238.0513 |
| M23 | 37.13 | [M-H]- | 349.0026 | 349.0024 | 0.7 | C15H10O8S | 350.01 | Apigenin+sulfation | 269.0456 |
| M24 | 37.13 | [M-H]- | 525.0368 | 525.0345 | 4.5 | C21H18O14S | 526.04 | Apigenin+glucuronidation+sulfation | 525.0361;349.0030;269.0451 |
| M25 | 39.94 | [M-H]- | 413.1091 | 413.1089 | 0.4 | C18H22O11 | 414.12 | Sinapine-( C4H10N+)+glucuronidation | 222.0531;207.0301 |
| M26 | 41.16 | [M-H]- | 287.0229 | 287.0231 | -0.7 | C11H12O7S | 288.03 | Sinapic acid+dehydroxylation+sulfation | / |
| M27 | 42.29 | [M-H]- | 445.0780 | 445.0776 | 0.8 | C21H18O11 | 446.08 | Apigenin+glucuronidation | 269.0446 |
| M28 | 43.18 | [M-H]- | 445.0784 | 445.0776 | 1.7 | C21H18O11 | 446.08 | Apigenin+glucuronidation | / |
| M29 | 50.98 | [M-H]- | 349.0020 | 349.0024 | -1.0 | C15H10O8S | 350.01 | Apigenin+sulfation | / |
| M30 | 51.45 | [M-H]- | 349.0027 | 349.0024 | 1.0 | C15H10O8S | 350.01 | Apigenin+sulfation | 269.0456117.0355 |
Table 2 Analysis and identification of metabolites in rat plasma of SZF based on UPLC-Q-TOF-MS
| Peak No. | Rt (min) | Selected- ion | Measured mass (m/z) | Ture values (m/z) | Error (ppm) | Formula | Molecular weight | Name | MS/MS |
|---|---|---|---|---|---|---|---|---|---|
| M1 | 4.14 | [M-H]- | 233.0120 | 233.0125 | 0.3 | C8H10O6S | 234.02 | Hydroxytyrosol+sulfation | 233.0120;153.0555;123.0449 |
| M2 | 4.54 | [M-H]- | 233.0125 | 233.0125 | 0.0 | C8H10O6S | 234.02 | Hydroxytyrosol+sulfation | 233.0109;153.0551;123.0447 |
| M3 | 5.70 | [M-H]- | 261.0081 | 261.0074 | 2.5 | C9H10O7S | 262.01 | Caffeic acid+hydrogenation+sulfation | / |
| M4 | 7.59 | [M-H]- | 247.0281 | 247.0282 | -0.3 | C9H12O6S | 248.04 | Hydroxytyrosol+methylation+sulfation | / |
| M5 | 8.44 | [M-H]- | 357.0828 | 357.0827 | 0.2 | C15H18O10 | 358.09 | Homovanillic acid+glucuronidation | 181.0494;175.0262;137.0605;113.0230 |
| M6 | 9.95 | [M-H]- | 245.0124 | 245.0125 | -0.5 | C9H10O6S | 246.02 | 4-Hydroxybenzoylcholine-(C3H8N+)+sulfation | 245.011;6165.0554;121.0651 |
| M7 | 11.51 | [M-H]- | 343.1034 | 343.1035 | -0.1 | C15H20O9 | 344.11 | Homovanillic alcohol+glucuronidation | 343.1300;167.0690;152.0490;113.0240 |
| M8 | 12.50 | [M-H]- | 275.0230 | 275.0231 | -0.4 | C10H12O7S | 276.03 | Genipinic acid+dehydroxylation+sulfation | / |
| M9 | 12.74 | [M-H]- | 371.0981 | 371.0984 | -0.7 | C16H20O10 | 372.11 | Genipinic acid+dehydroxylation+glucuronidation | 177.0519 |
| M10 | 13.62 | [M-H]- | 258.9917 | 258.9918 | -0.4 | C9H8O7S | 260.00 | Caffeic acid+sulfation | / |
| M11 | 13.73 | [M-H]- | 275.0229 | 275.0231 | -0.7 | C10H12O7S | 276.03 | Dihydroferulic acid+sulfation | 275.0255;195.0689;136.0514 |
| M12 | 15.23 | [M-H]- | 327.0721 | 327.0722 | -0.2 | C14H16O9 | 328.08 | 4-Hydroxybenzoylcholine-(C4H10N+)+glucuronidation | 151.0396;136.0164;113.0242 |
| M13 | 16.14 | [M-H]- | 245.0124 | 245.0125 | -0.5 | C9H10O6S | 246.02 | 4-Hydroxybenzoylcholine-(C3H8N+)+sulfation | 165.0550;150.0330 |
| M14 | 17.29 | [M-H]- | 303.0179 | 303.0180 | -0.4 | C11H12O8S | 304.03 | Sinapic acid+sulfation | / |
| M15 | 17.29 | [M-H]- | 303.0182 | 303.0180 | 0.6 | C11H12O8S | 304.03 | Sinapic acid+sulfation | / |
| M16 | 18.23 | [M-H]- | 273.0065 | 273.0074 | -3.5 | C10H10O7S | 274.01 | Ferulic acid+sulfation | 193.0505;178.0263;134.0368 |
| M17 | 19.53 | [M-H]- | 399.0930 | 399.0933 | -0.7 | C17H20O11 | 400.10 | Sinapic acid+glucuronidation | / |
| M18 | 19.62 | [M-H]- | 273.0077 | 273.0074 | 0.9 | C10H10O7S | 274.01 | Caffeic acid+methylation+sulfation | / |
| M19 | 20.10 | [M-H]- | 273.0075 | 273.0074 | 0.2 | C10H10O7S | 274.01 | Ferulic acid+sulfation | 193.0508;178.0275;134.0382 |
| M20 | 20.69 | [M-H]- | 399.0967 | 399.0933 | 8.6 | C17H20O11 | 400.10 | Sinapic acid+glucuronidation | / |
| M21 | 29.35 | [M-H]- | 621.1120 | 621.1097 | 3.7 | C27H26O17 | 622.12 | Apigenin+diglucuronidation | / |
| M22 | 34.77 | [M-H]- | 539.0508 | 539.0501 | 1.3 | C22H20O14S | 540.06 | Apigenin+methylation+glucuronidation+sulfation | 363.0188;283.0599;238.0513 |
| M23 | 37.13 | [M-H]- | 349.0026 | 349.0024 | 0.7 | C15H10O8S | 350.01 | Apigenin+sulfation | 269.0456 |
| M24 | 37.13 | [M-H]- | 525.0368 | 525.0345 | 4.5 | C21H18O14S | 526.04 | Apigenin+glucuronidation+sulfation | 525.0361;349.0030;269.0451 |
| M25 | 39.94 | [M-H]- | 413.1091 | 413.1089 | 0.4 | C18H22O11 | 414.12 | Sinapine-( C4H10N+)+glucuronidation | 222.0531;207.0301 |
| M26 | 41.16 | [M-H]- | 287.0229 | 287.0231 | -0.7 | C11H12O7S | 288.03 | Sinapic acid+dehydroxylation+sulfation | / |
| M27 | 42.29 | [M-H]- | 445.0780 | 445.0776 | 0.8 | C21H18O11 | 446.08 | Apigenin+glucuronidation | 269.0446 |
| M28 | 43.18 | [M-H]- | 445.0784 | 445.0776 | 1.7 | C21H18O11 | 446.08 | Apigenin+glucuronidation | / |
| M29 | 50.98 | [M-H]- | 349.0020 | 349.0024 | -1.0 | C15H10O8S | 350.01 | Apigenin+sulfation | / |
| M30 | 51.45 | [M-H]- | 349.0027 | 349.0024 | 1.0 | C15H10O8S | 350.01 | Apigenin+sulfation | 269.0456117.0355 |

Figure 1 Network pharmacology analysis A: venn diagram of target genes in treating UAN with SZF; B: PPI of differential expression genes in treating UAN with SZF; C: enrichment analysis of GO function; D: enrichment analysis of KEGG pathway. SZF: Shizhi Fang; UAN: uric acid nephropathy; PPI: Protein-Protein Interaction; GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes.
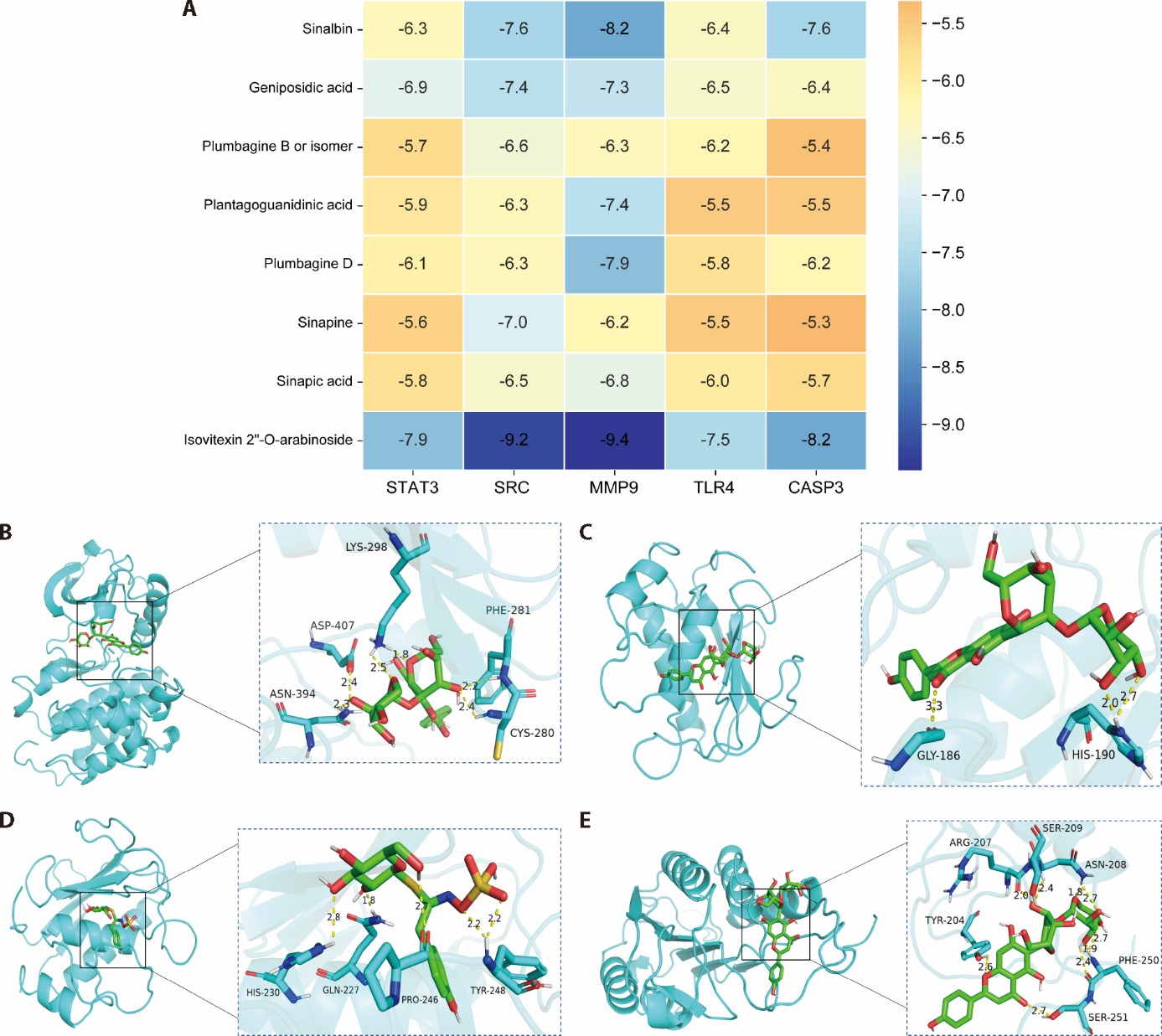
Figure 2 Molecular docking diagram A: molecular docking binding energy; B: isovitexin 2-O-arabinoside-SRC; C: isovitexin 2-O-arabinoside-MMP9; D: Sinalbin-MMP9; E: isovitexin 2-O-arabinoside-CASP3. SRC: SRC proto-oncogene, non-receptor tyrosine kinase; MMP9: matrix metallopeptidase 9; CASP3: Caspase3.
| Group | n | Creatinine (μmol/L) | Blood urea nitrogen (mmol/L) | Serum uric acid (μmol/L) | Urinary uric acid (μmol/L) | Urinary total protein (mg/L) | Urine albumin-to-creatinine ratio (mg/g) |
|---|---|---|---|---|---|---|---|
| Normal | 6 | 12.00±2.44 | 8.68±0.51 | 87.33±21.47 | 532.80±311.60 | 7.03±4.00 | 20.59±11.93 |
| Model | 6 | 16.50±2.25a | 11.60±1.38a | 182.20±14.47c | 212.20±60.00a | 43.47±12.99c | 133.2±82.32c |
| 0.234 g/kg | 6 | 14.00±4.00 | 11.00±3.28 | 137.20±45.76d | 429.80±262.00 | 24.18±28.35 | 32.56±34.39e |
| 0.468 g/kg | 6 | 13.00±2.44 | 9.93±0.59 | 137.20±28.19d | 461.20±240.60 | 6.72±5.32e | 19.85±13.45e |
| 0.936 g/kg | 6 | 11.17±1.72b | 8.46±0.94b | 97.83±21.40e | 582.80±151.20d | 7.78±4.64e | 15.19±11.73e |
| Febuxostat | 6 | 11.00±0.63b | 8.16±0.19b | 81.00±5.69e | 540.80±107.20d | 5.24±1.72e | 16.13±8.27e |
Table 3 Biochemical indicators of each group of mice ($\bar{x}$ ± s)
| Group | n | Creatinine (μmol/L) | Blood urea nitrogen (mmol/L) | Serum uric acid (μmol/L) | Urinary uric acid (μmol/L) | Urinary total protein (mg/L) | Urine albumin-to-creatinine ratio (mg/g) |
|---|---|---|---|---|---|---|---|
| Normal | 6 | 12.00±2.44 | 8.68±0.51 | 87.33±21.47 | 532.80±311.60 | 7.03±4.00 | 20.59±11.93 |
| Model | 6 | 16.50±2.25a | 11.60±1.38a | 182.20±14.47c | 212.20±60.00a | 43.47±12.99c | 133.2±82.32c |
| 0.234 g/kg | 6 | 14.00±4.00 | 11.00±3.28 | 137.20±45.76d | 429.80±262.00 | 24.18±28.35 | 32.56±34.39e |
| 0.468 g/kg | 6 | 13.00±2.44 | 9.93±0.59 | 137.20±28.19d | 461.20±240.60 | 6.72±5.32e | 19.85±13.45e |
| 0.936 g/kg | 6 | 11.17±1.72b | 8.46±0.94b | 97.83±21.40e | 582.80±151.20d | 7.78±4.64e | 15.19±11.73e |
| Febuxostat | 6 | 11.00±0.63b | 8.16±0.19b | 81.00±5.69e | 540.80±107.20d | 5.24±1.72e | 16.13±8.27e |
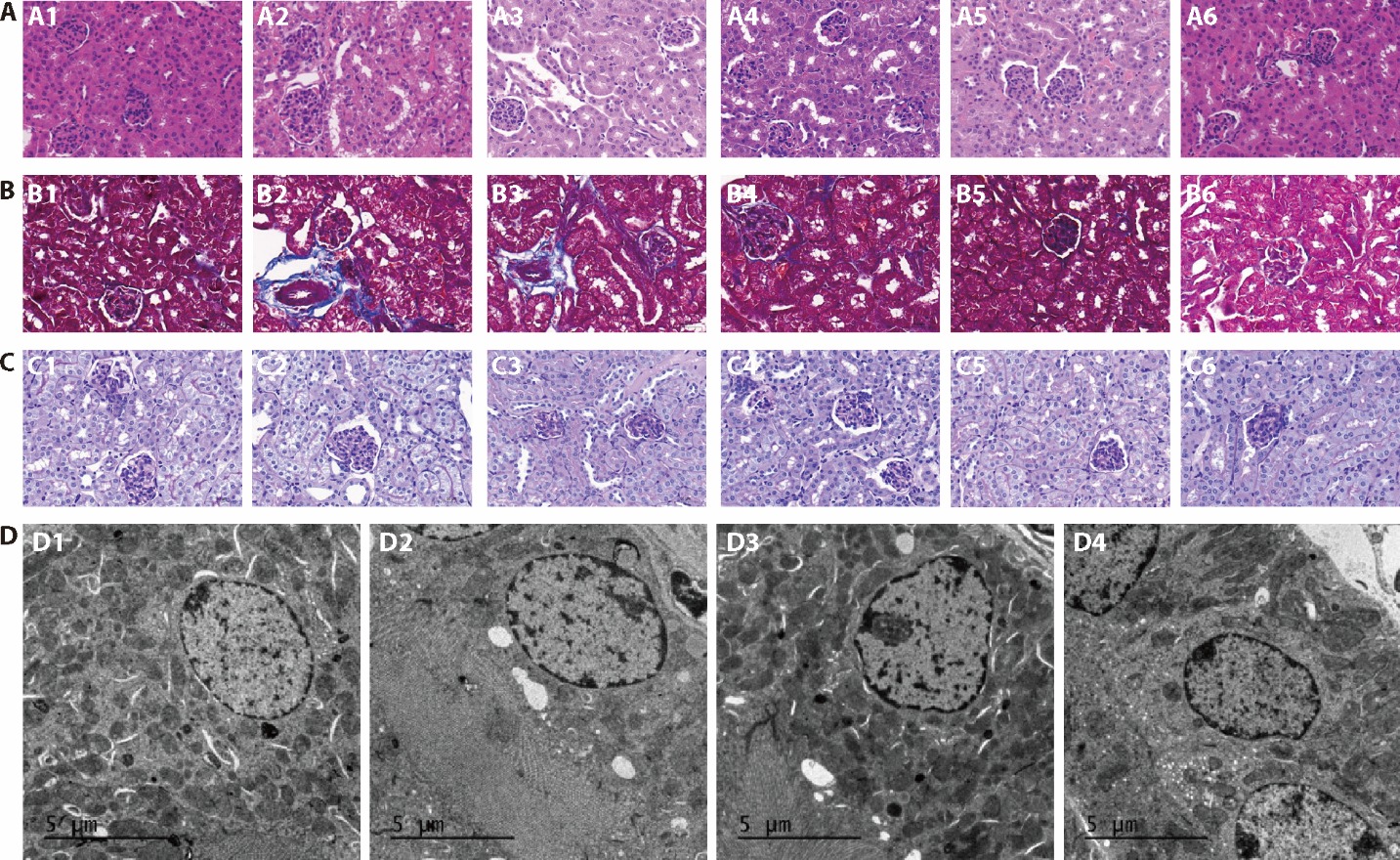
Figure 3 Renal pathology of mice kidney tissue in groups A: HE staining, B: Masson staining; C: PAS staining (× 200, 50 μm); D: TEM of mice kidney tissue in groups (× 4200, 5 μm). A1, B1, C1, D1: normal group; A2, B2, C2, D2: model group; A3, B3, C3: 0.234 g/kg group; A4, B4, C4: 0.468 g/kg group; A5, B5, C5, D3: 0.936 g/kg group; A6, B6, C6, D4: febuxostat group. Normal group: without hyperuricemia model; model group: without any treatment; 0.234 g/kg group: treated with 0.234 g/kg SZF for 4 weeks; 0.468 g/kg group: treated with 0.468 g/kg SZF for 4 weeks; 0.936 g/kg group: treated with 0.936 g/kg SZF for 4 weeks; Febuxostat group: treated with 6 mg/kg febuxostat for 4 weeks. SZF: Shizhi Fang; HE: hematoxylin and eosin staining; PAS: periodic acid-schiff staining; TEM: transmission electron microscopy.
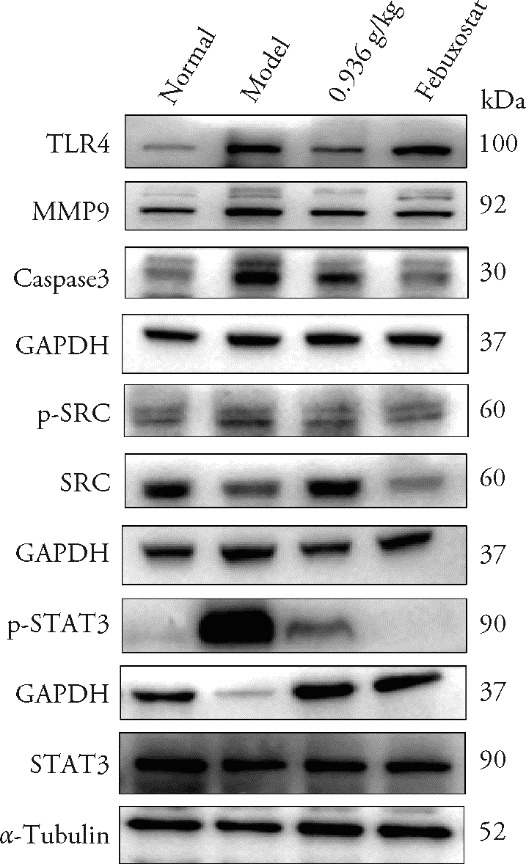
Figure 4 Expression of TLR4, MMP9, Caspase-3, p-SRC, SRC, p-STAT3 and STAT3 proteins in renal tissue Normal group: without hyperuricemia model; model group: without any treatment; 0.936 g/kg group: treated with 0.936 g/kg SZF for 4 weeks; Febuxostat group: treated with 6 mg/kg febuxostat for 4 weeks. SZF: Shizhi Fang; TLR4: toll-like receptor 4; MMP9: matrix metalloproteinase-9; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; p-SRC: phos-phorylated proto-oncogene tyrosine-protein kinase Src; SRC: proto-oncogene tyrosine-protein kinase Src; p-STAT3: phos-phorylated signal transducer and activator of transcription 3; STAT3: signal transducer and activator of transcription 3.
| Group | n | Caspase3 /GAPDH | MMP9 /GAPDH | TLR4 /GAPDH | p-SRC /SRC | p-STAT3 /STAT3 |
|---|---|---|---|---|---|---|
| Normal | 3 | 0.580±0.010 | 0.720±0.010 | 0.190±0.010 | 0.880±0.020 | 0.080±0.020 |
| Model | 3 | 0.980±0.010a | 1.140±0.000a | 0.980±0.000a | 1.120±0.030a | 1.190±0.020a |
| 0.936 g/kg | 3 | 0.700±0.010b | 0.750±0.000b | 0.560±0.010b | 0.740±0.040b | 0.520±0.030b |
| Febuxostat | 3 | 0.610±0.020b | 0.750±0.000b | 0.870±0.010b | 0.410±0.050b | 0.070±0.050b |
Table 4 Expression of TLR4, MMP9, Caspase-3, p-SRC, SRC, p-STAT3 and STAT3 proteins in renal tissue ($\bar{x}$ ± s)
| Group | n | Caspase3 /GAPDH | MMP9 /GAPDH | TLR4 /GAPDH | p-SRC /SRC | p-STAT3 /STAT3 |
|---|---|---|---|---|---|---|
| Normal | 3 | 0.580±0.010 | 0.720±0.010 | 0.190±0.010 | 0.880±0.020 | 0.080±0.020 |
| Model | 3 | 0.980±0.010a | 1.140±0.000a | 0.980±0.000a | 1.120±0.030a | 1.190±0.020a |
| 0.936 g/kg | 3 | 0.700±0.010b | 0.750±0.000b | 0.560±0.010b | 0.740±0.040b | 0.520±0.030b |
| Febuxostat | 3 | 0.610±0.020b | 0.750±0.000b | 0.870±0.010b | 0.410±0.050b | 0.070±0.050b |
| 1. |
Kang DH, Nakagawa T, Feng L, et al. A role for uric acid in the progression of renal disease. J Am Soc Nephrol 2002; 13: 2888-97.
DOI URL |
| 2. |
Li Y, Shen Z, Zhu B, Zhang H, Zhang X, Ding X. Demographic, regional and temporal trends of hyperuricemia epidemics in mainland China from 2000 to 2019: a systematic review and Meta-analysis. Glob Health Action 2021; 14: 1874652.
DOI URL |
| 3. |
Zhang M, Zhu X, Wu J, et al. Prevalence of hyperuricemia among chinese adults: findings from two nationally representative Cross-sectional surveys in 2015-16 and 2018-19. Front Immunol 2021; 12: 791983.
DOI URL |
| 4. |
Dalbeth N, Choi HK, Joosten LAB, et al. Gout. Nat Rev Dis Primers 2019; 5: 69.
DOI PMID |
| 5. |
Narang RK, Dalbeth N. Pathophysiology of gout. Semin Nephrol 2020; 40: 550-63.
DOI PMID |
| 6. |
Diaz-Torne C, Ortiz MA, Garcia-Guillen A, et al. The inflammatory role of silent urate crystal deposition in intercritical gout. Rheumatology (Oxford) 2021; 60: 5463-72.
DOI PMID |
| 7. |
Sun X, Yang L, Sun H, et al. TCM and related active compounds in the treatment of gout: the regulation of signaling pathway and urate transporter. Front Pharmacol 2023; 14: 1275974.
DOI URL |
| 8. |
Han R, Qiu H, Zhong J, et al. Si Miao Formula attenuates non-alcoholic fatty liver disease by modulating hepatic lipid metabolism and gut microbiota. Phytomedicine 2021; 85: 153544.
DOI URL |
| 9. |
Zhou H, Yang J, Yuan X, et al. Hyperuricemia research progress in model construction and Traditional Chinese Medicine interventions. Front Pharmacol 2024; 15: 1294755.
DOI URL |
| 10. |
Li S, Zhang B. Traditional Chinese Medicine network pharmacology: theory, methodology and application. Chin J Nat Med 2013; 11: 110-20.
DOI URL |
| 11. | Zhang P, Zhang D, Zhou W, et al. Network pharmacology: towards the artificial intelligence-based precision Traditional Chinese Medicine. Brief Bioinform 2023; 25: bbad518. |
| 12. | Wang Z, Wang X, Zhang D, Hu Y, Li S. Traditional Chinese Medicine network pharmacology: development in new era under guidance of network pharmacology evaluation method guidance. Zhong Guo Zhong Yao Za Zhi 2022; 47: 7-17. |
| 13. | Li Y, He L. Treatment of 33 cases of hyperuricemic nephropathy of phlegm-blood stasis by Shizhi decoction. Shanghai Zhong Yi Yao Za Zhi 2011; 45: 41-3. |
| 14. | Wang L. The effect of Shizhi decoction on oxidative stress in elderly patients with uric acid nephropathy. Zhong Guo Lao Nian Xue Za Zhi 2016; 36: 4556-8. |
| 15. | Szklarczyk D, Kirsch R, Koutrouli M, et al. The STRING database in 2023:protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res 2023; 51: D638-46. |
| 16. |
Sherman BT, Hao M, Qiu J, et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res 2022; 50: W216-21.
DOI PMID |
| 17. | Wu Z, Wang C, Yang F, et al. Network pharmacology, molecular docking, combined with experimental verification to explore the role and mechanism of shizhifang decoction in the treatment of hyperuricemia. Heliyon 2024; 10: e24865. |
| 18. | Zhou J, Wang C, Zhang X, et al. Shizhi Fang ameliorates pyroptosis of renal tubular epithelial cells in hyperuricemia through inhibiting NLRP 3 inflammasome. J Ethnopharmacol 2023; 317: 116777. |
| 19. |
Li D, Li Y, Chen X, et al. The pathogenic mechanism of monosodium urate crystal-induced kidney injury in a rat model. Front Endocrinol (Lausanne) 2024; 15: 1416996.
DOI URL |
| 20. |
Liu T, Gao H, Zhang Y, et al. Apigenin ameliorates hyperuricemia and renal injury through regulation of uric acid metabolism and JAK2/STAT3 signaling pathway. Pharmaceuticals (Basel) 2022; 15: 1442.
DOI URL |
| 21. |
Lu M, Yin J, Xu T, et al. Fuling-Zexie formula attenuates hyperuricemia-induced nephropathy and inhibits JAK2/STAT3 signaling and NLRP 3 inflammasome activation in mice. J Ethnopharmacol 2024; 319: 117262.
DOI URL |
| 22. |
Zhang Y, Wang S, Dai X, et al. Simiao San alleviates hyperuricemia and kidney inflammation by inhibiting NLRP 3 inflammasome and JAK2/STAT3 signaling in hyperuricemia mice. J Ethnopharmacol 2023; 312: 116530.
DOI URL |
| 23. |
Xiong C, Deng J, Wang X, Hou Q, Zhuang S. Pharmacological inhibition of Src family kinases attenuates hyperuricemic nephropathy. Front Pharmacol 2024; 15: 1352730.
DOI URL |
| 24. |
Pan J, Shi M, Li L, et al. Pterostilbene, a bioactive component of blueberries, alleviates renal fibrosis in a severe mouse model of hyperuricemic nephropathy. Biomed Pharmacother 2019; 109: 1802-8.
DOI PMID |
| 25. | Xiao J, Zhang X, Fu C, et al. Impaired Na(+)-K(+)-ATPase signaling in renal proximal tubule contributes to hyperuricemia-induced renal tubular injury. Exp Mol Med 2018; 50: e452. |
| 26. |
Yu W, Huang G, Wang J, et al. Imperata cylindrica polysaccharide ameliorates intestinal dysbiosis and damage in hyperuricemic nephropathy. Int J Biol Macromol 2024; 278: 134432.
DOI URL |
| 27. | Fan W, Liu C, Chen D, et al. Ozone alleviates MSU-induced acute gout pain via upregulating AMPK/GAS6/MerTK/SOCS 3 signaling pathway. J Transl Med 2023; 21: 890. |
| 28. |
Miao Z, Guo W, Lu S, et al. Hypothermia induced by adenosine 5'-monophosphate attenuates early stage injury in an acute gouty arthritis rat model. Rheumatol Int 2013; 33: 2085-92.
DOI PMID |
| 29. | Wang J, Tsai S, Tsai H, Lin S, Huang P. Hyperuricemia exacerbates abdominal aortic aneurysm formation through the URAT1/ERK/MMP-9 signaling pathway. BMC Cardiovasc Disord 2023; 23: 55. |
| 30. |
Liu Y, Han Y, Liu Y, et al. Xanthoceras sorbifolium leaves alleviate hyperuricemic nephropathy by inhibiting the PI3K/AKT signaling pathway to regulate uric acid transport. J Ethnopharmacol 2024; 327: 117946.
DOI URL |
| 31. | Tao M, Shi Y, Tang L, et al. Blockade of ERK1/2 by U0126 alleviates uric acid-induced EMT and tubular cell injury in rats with hyperuricemic nephropathy. Am J Physiol Renal Physiol 2019; 316: F660-F73. |
| 32. | Huang Z, Xie N, Illes P, et al. From purines to purinergic signalling: molecular functions and human diseases. Signal Transduct Target Ther 2021; 6: 162. |
| 33. |
Balakumar P, Alqahtani A, Khan NA, Mahadevan N, Dhanaraj SA. Mechanistic insights into hyperuricemia-associated renal abnormalities with special emphasis on epithelial-to-mesenchymal transition: pathologic implications and putative pharmacologic targets. Pharmacol Res 2020; 161: 105209.
DOI URL |
| 34. | Crisan TO, Cleophas MC, Oosting M, et al. Soluble uric acid primes TLR-induced proinflammatory cytokine production by human primary cells via inhibition of IL-1Ra. Ann Rheum Dis 2016; 75: 755-62. |
| 35. |
Liu H, Chen Z, Liu M, et al. The Terminalia chebula Retz extract treats hyperuricemic nephropathy by inhibiting TLR4/ MyD88/NF-kappa B axis. J Ethnopharmacol 2024; 322: 117678.
DOI URL |
| 36. |
Yang Y, Zhang D, Liu J, et al. Wuling San protects kidney dysfunction by inhibiting renal TLR4/MyD88 signaling and NLRP3 inflammasome activation in high fructose-induced hyperuricemic mice. J Ethnopharmacol 2015; 169: 49-59.
DOI PMID |
| 37. | Ouyang X, Li N, Guo M, et al. Active flavonoids from lagotis brachystachya attenuate monosodium urate-induced gouty arthritis via inhibiting TLR4/MyD88/NF-kappaB pathway and NLRP 3 expression. Front Pharmacol 2021; 12: 760331. |
| 38. | Ishaq M, Zhao L, Soliman MM, et al. Ameliorative impacts of Sinapic acid against monosodium urate crystal-induced gouty arthritis and inflammation through different signaling pathways. Toxicol Res (Camb) 2024; 13: tfae130. |
| 39. | Ishaq M, Mehmood A, Ur Rehman A, et al. Antihyperuricemic effect of dietary polyphenol sinapic acid commonly present in various edible food plants. J Food Biochem 2020; 44: e13111. |
| 40. | Wang K, Liang C, Cao W, et al. Dietary sinapic acid attenuated high-fat diet-induced lipid metabolism and oxidative stress in male Syrian hamsters. J Food Biochem 2022; 46: e14203. |
| 41. |
Boulghobra D, Grillet PE, Laguerre M, et al. Sinapine, but not sinapic acid, counteracts mitochondrial oxidative stress in cardiomyocytes. Redox Biol 2020; 34: 101554.
DOI URL |
| 42. | Li Y, Xu YJ, Tan CP, Liu Y. Sinapine improves LPS-induced oxidative stress in hepatocytes by down-regulating MCJ protein expression. Life Sci 2022; 306: 120828. |
| 43. | Guo Y, Ding Y, Zhang T, An H. Sinapine reverses multi-drug resistance in MCF-7/dox cancer cells by downregulating FGFR4/FRS2alpha-ERK1/2 pathway-mediated NF-kappaB activation. Phytomedicine 2016; 23: 267-73. |
| 44. |
Zheng X, Mao C, Qiao H, et al. Plumbagin suppresses chronic periodontitis in rats via down-regulation of TNF-alpha, IL-1beta and IL-6 expression. Acta Pharmacol Sin 2017; 38: 1150-60.
DOI URL |
| 45. |
Wang Y, Li X, Wang W, Zou L, Miao H, Zhao Y. Geniposidic acid attenuates chronic tubulointerstitial nephropathy through regulation of the NF-kB/Nrf2 pathway via aryl hydrocarbon receptor signaling. Phytother Res 2024; 38: 5441-57.
DOI URL |
| 46. |
Guo Y, Bao C, Ma D, et al. Network-Based combinatorial CRISPR-Cas9 screens identify synergistic modules in Human cells. ACS Synth Biol 2019; 8: 482-90.
DOI PMID |
| 47. |
Gan C, Tao QW, Yi HY, et al. Network Meta-analysis of the clinical efficacy and safety of kidney-tonifying and bone-strengthening therapies for the treatment of rheumatoid arthritis with kidney deficiency type. J Tradit Chin Med 2024; 44: 1067-81.
DOI |
| [1] | ZHANG Yuan, CHENG Shizan, HUA Yue, SHI Ji, SU Guoming, ZHANG Chao, LIAN Jing, LIU Pengpeng, JIA Tianzhu. Mechanisms of Suanzaoren (Ziziphi Spinosae Semen) and its processed products in treating insomnia: an integrated study based on network pharmacology and metabolomics [J]. Journal of Traditional Chinese Medicine, 2025, 45(6): 1307-1316. |
| [2] | LI Keyao, SHU Ye, CHANG Jing, TANG Jianping, ZHANG Litao, WEI Zhu. Psoriasis intervention by Huai’er (Trametes): unveiling novel targets via network pharmacology [J]. Journal of Traditional Chinese Medicine, 2025, 45(6): 1317-1329. |
| [3] | MA Guiping, CHEN Ran, LI Junlong, SUN Le, HU Shiping, ZHANG Yiyi, HONG Chuangxiong. Network pharmacology combined with in vivo experiments to explore the molecular mechanism of Jiawei Erzhi pill (加味二至丸) protects against atherosclerosis by inhibiting ferroptosis [J]. Journal of Traditional Chinese Medicine, 2025, 45(6): 1330-1341. |
| [4] | ZHAO Yumin, ZHANG Yuliang, WANG Guozi, LIU Xizan, ZHAO Pengmin, ZHAO Mengjun, LI Zhaoxia, DI Haixia. Network pharmacology-based analysis of the antithrombotic clinical efficacy and antithrombotic mechanism of Huoxue Jiedu prescription (活血解毒方) in the treatment of polycythemia vera with heat toxin and blood stasis syndrome [J]. Journal of Traditional Chinese Medicine, 2025, 45(6): 1353-1365. |
| [5] | PEI Ke, LI Yong, LIN Zhe, LYU Guangfu. Mechanisms of Baishao (Radix Paeoniae Alba) and Gancao (Radix Glycyrrhizae) on major depressive disorder: network pharmacology and in vivo validation [J]. Journal of Traditional Chinese Medicine, 2025, 45(5): 1067-1077. |
| [6] | ZHANG Wei, REN Changhong, GAO Chen, XU Jun, WU Xiaodan, YANG Yong. Systematic understanding of mechanism of Shenfu decoction (参附汤) improve the prognosis of ischemic stroke using a network pharmacology and animal experiment approach [J]. Journal of Traditional Chinese Medicine, 2025, 45(5): 1078-1086. |
| [7] | QI Yafeng, LIU Yu, LIU Yeyuan, LI Yangyang, ZHANG Shangzu, CHEN Yaping, XU Qian, HAO Guoxiong, LIU Yongqi, ZHANG Liying, ZHANG Zhiming. Therapeutic potential of Traditional Chinese Medicine Yisui Shengxue pills (益髓生血丸) to inhibit hypoxia-inducible factor-1alpha and general control nonderepressible 2 to regulate the post-chemotherapy immune response: integrating network pharmacology and experimental validation [J]. Journal of Traditional Chinese Medicine, 2025, 45(5): 1087-1097. |
| [8] | CHOI You Yeon, JIN Seong chul, KIM Mi Hye, BAEK Hee Kyung, KIM Dong Hyun, OH Sung Hyuk, YANG Woong Mo. Exploring the therapeutic potential of Morus alba Linne extract in targeting localized adiposity [J]. Journal of Traditional Chinese Medicine, 2025, 45(5): 970-978. |
| [9] | SONG Mingming, MEN Bo, CHEN Mei, LIU Rui, MO Hongping, ZHANG Da, PAN Tao, WEN Xudong. Exploration of the mechanism of Danggui Buxue decoction (当归补血汤) for the treatment of gastric ulcer based on network pharmacology, molecular docking, and in vivo experiment [J]. Journal of Traditional Chinese Medicine, 2025, 45(4): 806-816. |
| [10] | LI Yue, DENG Jinyan, PI Shanshan, ZHANG Yingjuan, ZHAO Dan, GUO Yi, YE Yong’an, ZAO Xiaobin, DU Hongbo. Weifuchun (胃复春 ) exerts therapeutic effects on gastric fundic gland polyps by promoting ferroptosis [J]. Journal of Traditional Chinese Medicine, 2025, 45(3): 618-627. |
| [11] | HUANG Jiaen, LUO Qing, DONG Gengting, PENG Weiwen, HE Jianhong, DAI Weibo. Xiahuo Pingwei San (夏藿平胃散) attenuated intestinal inflammation in dextran sulfate sodium-induced ulcerative colitis mice through inhibiting the receptor for advanced glycation end-products signaling pathway [J]. Journal of Traditional Chinese Medicine, 2025, 45(2): 311-325. |
| [12] | HAN Shuai, Du Zhikang, WANG Zirui, HUANG Tianfeng, GE Yali, SHI Jianwen, GAO Ju. Network pharmacology approach to unveiling the mechanism of berberine in the amelioration of morphine tolerance [J]. Journal of Traditional Chinese Medicine, 2025, 45(2): 376-384. |
| [13] | TAN Xiying, GU Ruxin, TAO Jing, ZHANG Yu, SUN RuiQian, YIN Gang, ZHANG Shuo, TANG Decai. Integrating network pharmacology and experimental validation to uncover the synergistic effects of Huangqi (Radix Astragali Mongolici)-Ezhu (Rhizoma Curcumae Phaeocaulis) with 5-fluorouracil in colorectal cancer models [J]. Journal of Traditional Chinese Medicine, 2025, 45(2): 385-398. |
| [14] | SHI Jinyu, PAN Fuwei, GE Haiya, YANG Zongrui, ZHAN Hongsheng. Mechanism of Qigu capsule (芪骨胶囊) as a treatment for sarcopenia based on network pharmacology and experimental validation [J]. Journal of Traditional Chinese Medicine, 2025, 45(2): 399-407. |
| [15] | HU Huiming, WENG Jiajun, TANG Fangrui, WANG Yaqi, FAN Shengxian, WANG Xuecheng, CUI Can, SHAO Feng, ZHU Yanchen. Hypolipidemic effect and mechanism of Hedan tablet (荷丹片) based on network pharmacology [J]. Journal of Traditional Chinese Medicine, 2025, 45(2): 408-421. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||

