Journal of Traditional Chinese Medicine ›› 2025, Vol. 45 ›› Issue (4): 909-921.DOI: 10.19852/j.cnki.jtcm.2025.04.021
• Original Articles • Previous Articles Next Articles
Identification of characteristic genes of Yin and Yang deficiency constitutions: an integrated analysis based on bioinformatics and machine learning
LONG Xi1,2, WU Zixuan1,2, YU Yunfeng1,2, LIN Jie3( ), PENG Qinghua2,4(
), PENG Qinghua2,4( )
)
- 1 Department of Graduate School, Hunan University of Chinese Medicine, Changsha 410208, China
2 Department of Chinese Medicine, Hunan University of Chinese Medicine, Changsha 410208, China
3 Department of Obstetrics and Gynecology, The First Affiliated Hospital of Hunan University of Chinese Medicine, Changsha 410007, China
4 Department of Ophthalmology, The First Affiliated Hospital of Hunan University of Chinese Medicine, Changsha 410007, China
-
Received:2024-12-22Accepted:2025-05-26Online:2025-08-15Published:2025-07-25 -
Contact:LIN Jie,PENG Qinghua -
About author:Prof. LIN Jie, Department of Obstetrics and Gynecology, the First Affiliated Hospital of Hunan University of Chinese Medicine, Changsha 410007, China. 978476646@qq.com.Telephone: +86-18274563185
Prof. PENG Qinghua, Department of Chinese Medicine, Hunan University of Chinese Medicine, Changsha 410208, China. pengqinghua@hnucm.edu.cn;
First author contact:LONG Xi and WU Zixuan are co-first authors and contributed equally to this work
-
Supported by:National Natural Science Foundation of China: Establishment and Validation of a High-throughput Screening System for Traditional Chinese Medicine Against Dry Eye Disease based on transforming growth factor-βand B-cell lymphoma-2(81574031);2023 Hunan Provincial Department of Education Scientific Research Project: Yiguanjian Decoction Regulates Th17/Treg Cell Balance and Its Intervention Mechanism in Dry Eye with Liver Yin Deficiency Syndrome(23A0300)
Cite this article
LONG Xi, WU Zixuan, YU Yunfeng, LIN Jie, PENG Qinghua. Identification of characteristic genes of Yin and Yang deficiency constitutions: an integrated analysis based on bioinformatics and machine learning[J]. Journal of Traditional Chinese Medicine, 2025, 45(4): 909-921.
share this article
| GEO accession | Title | Platform | Sample size | Sex information | Age (years) |
|---|---|---|---|---|---|
| GSE87474 | Expression Profiling-based Clustering of Healthy Subjects Recapitulates Classifications Defined by Clinical Observation in Chinese Medicine | GPL570 | 32 Chinese Han individuals with Yang/Yin deficiency (n = 12 each) and Balanced constitutions (n = 8) | 3 males for Yang/Yin deficiency and 9 females for Yang/Yin deficiency | 22.1 ± 2.4 for Yin deficiency and 23.6 ± 3.6 for Yang deficiency |
| GSE56116 | Genes expression profiles of postmenopausal osteoporosis with kidney Yin deficiency | GPL4133 | 10 Chinese with kidney Yin deficiency (n = 4), kidney Yang deficiency (n = 3) and, non-kidney deficiency (n = 3) | 0 males for Yang/Yin deficiency and 3/4 females for Yang/Yin deficiency | 60.5 ± 2.6 for Yin deficiency and 58.6 ± 6.1 for Yang deficiency |
Table 1 Specific information on the source of the data
| GEO accession | Title | Platform | Sample size | Sex information | Age (years) |
|---|---|---|---|---|---|
| GSE87474 | Expression Profiling-based Clustering of Healthy Subjects Recapitulates Classifications Defined by Clinical Observation in Chinese Medicine | GPL570 | 32 Chinese Han individuals with Yang/Yin deficiency (n = 12 each) and Balanced constitutions (n = 8) | 3 males for Yang/Yin deficiency and 9 females for Yang/Yin deficiency | 22.1 ± 2.4 for Yin deficiency and 23.6 ± 3.6 for Yang deficiency |
| GSE56116 | Genes expression profiles of postmenopausal osteoporosis with kidney Yin deficiency | GPL4133 | 10 Chinese with kidney Yin deficiency (n = 4), kidney Yang deficiency (n = 3) and, non-kidney deficiency (n = 3) | 0 males for Yang/Yin deficiency and 3/4 females for Yang/Yin deficiency | 60.5 ± 2.6 for Yin deficiency and 58.6 ± 6.1 for Yang deficiency |
| Item | Yin deficiency | Yang deficiency |
|---|---|---|
| Main characteristics | Heat intolerance, prefer cold food and drinks, susceptible to heat | Cold intolerance, cold hands, feet, stomach, and waist, prefer hot food and drinks, susceptible to cold |
| Secondary characteristics | Constipated, emaciation, red lips and dryness, thirst, insomnia | Watery stool, fat, whitish skin, nocturia, fluid retention, chills |
| Tongue and Pulse | Prefer red and dry tongue, prefer rapid pulse | Prefer pale and tender tongue, prefer slow pulse |
Table 2 Clinical symptom of Yin and Yang deficiency
| Item | Yin deficiency | Yang deficiency |
|---|---|---|
| Main characteristics | Heat intolerance, prefer cold food and drinks, susceptible to heat | Cold intolerance, cold hands, feet, stomach, and waist, prefer hot food and drinks, susceptible to cold |
| Secondary characteristics | Constipated, emaciation, red lips and dryness, thirst, insomnia | Watery stool, fat, whitish skin, nocturia, fluid retention, chills |
| Tongue and Pulse | Prefer red and dry tongue, prefer rapid pulse | Prefer pale and tender tongue, prefer slow pulse |

Figure 1 Differentially expressed genes analysis A: differentially expressed genes analysis in heatmap of DEGs in Yin deficiency; B: differentially expressed genes analysis in heatmap of DEGs in Yang deficiency. Display only the top 100 genes with differential rankings. DEGs: differentially expressed genes analysis.

Figure 2 Weighted gene co-expression network analysis A: selection of soft thresholds (β) for WGCNA analysis of Yin deficiency; A1: mean connectivity in Yin deficiency; A2: scale free topology model fit in Yin deficiency, signed R2; B: selection of soft thresholds (β) for WGCNA analysis of Yang deficiency; B1: mean connectivity in Yang deficiency; B2: scale free topology model fit in Yang deficiency, signed R2; C: this diagram shows the degree of correlation within the module of the Yin deficiency; D: this diagram shows the degree of correlation within the module of the Yang deficiency; E: relationship between modules and clinical features in Yin deficiency; F: relationship between modules and clinical features in Yang deficiency. WGCNA: weighted gene co-expression network analysis.
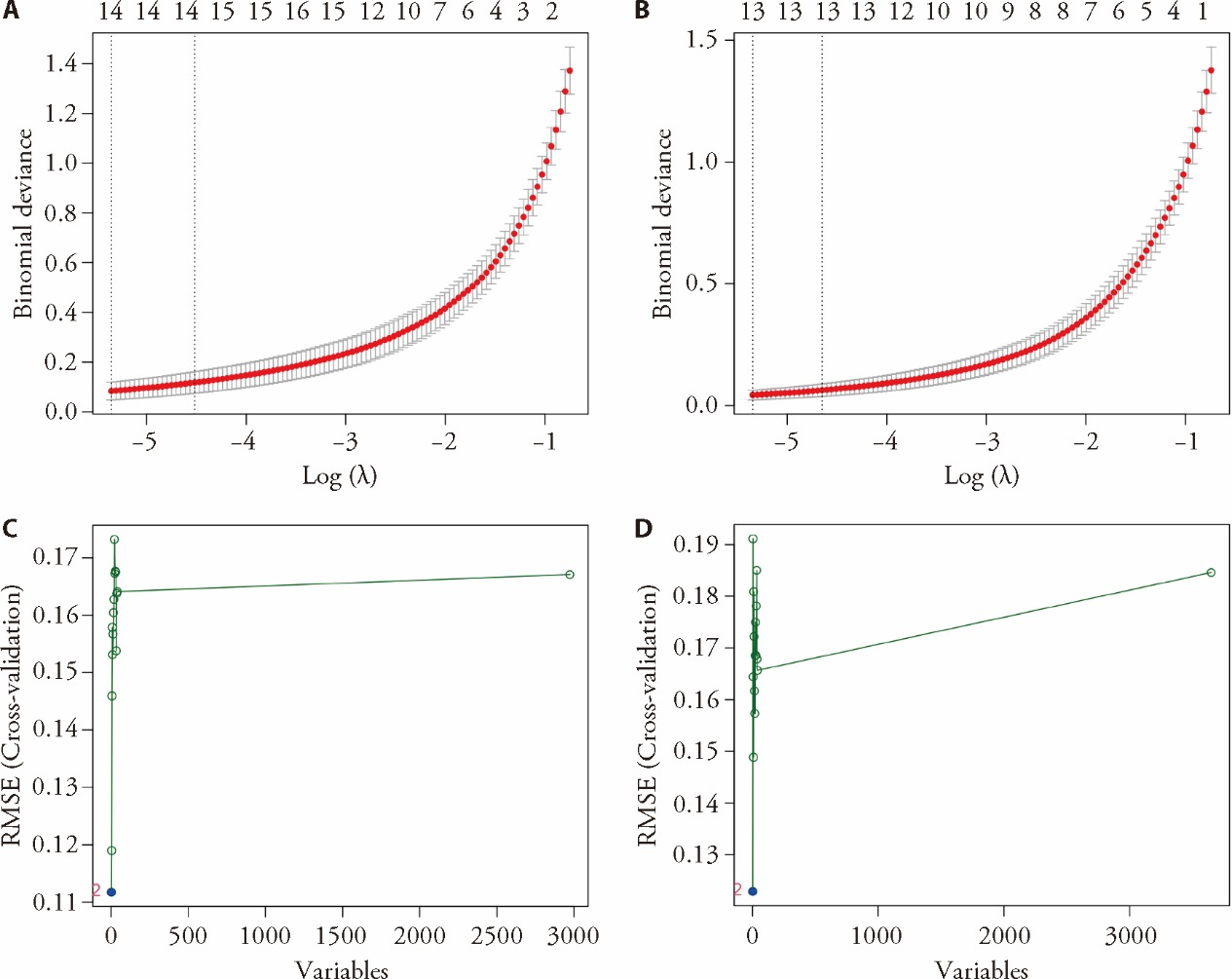
Figure 3 Machine learning analysis A: regression of the hub genes of Yin deficiency using LASSO; B: regression of the hub genes of Yang deficiency using LASSO; C: regression of the hub genes of Yin deficiency using SVM-RFE; D: regression of the hub genes of Yang deficiency using SVM-RFE. LASSO: least absolute shrinkage and selection operator; SVM-RFE: support vector machine-recursive feature elimination
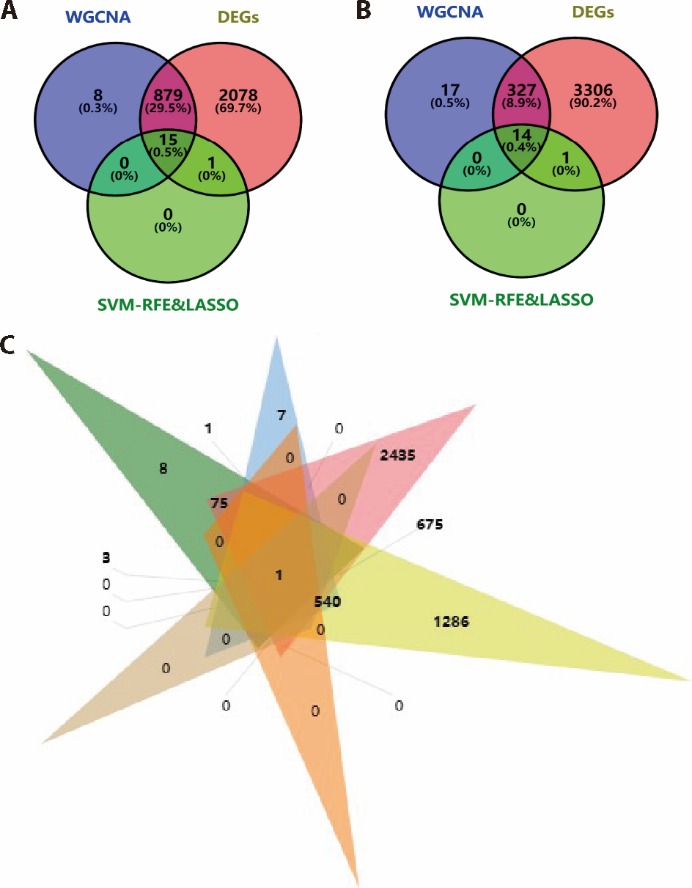
Figure 4 Hub genes indentification A: intersecting genes in Yin deficiency of DEGs, WGCNA and machine learning analysis. B: intersecting genes in Yang deficiency of DEGs, WGCNA and machine learning analysis. C: intersecting of hub genes in Yin deficiency and hub genes in Yang deficiency. DEGs: differentially expressed genes analysis; WGCNA: weighted gene co-expression network analysis; LASSO: least absolute shrinkage and selection operator; SVM-RFE: support vector machine-recursive feature elimination.
| 1. |
Sweeney EL, Bradshaw CS, Murray GL, Whiley DM. Individualised treatment of mycoplasma genitalium infection-incorporation of fluoroquinolone resistance testing into clinical care. Lancet Infect Dis 2022; 22: e267-70.
DOI PMID |
| 2. | Lyu W. Understanding Traditional Chinese Medicine. Hepatobiliary Surg Nutr 2021; 10: 846-8. |
| 3. | Wang J, Wong YK, Liao F. What has Traditional Chinese Medicine delivered for modern medicine? Expert Rev Mol Med 2018; 20: e4. |
| 4. | Yan Q. Stress and systemic inflammation: Yin-Yang dynamics in health and diseases. Methods Mol Biol 2018; 1781: 3-20. |
| 5. | Chan H, Ng T. Traditional Chinese Medicine (TCM) and allergic diseases. Curr Allergy Asthma Rep 2020; 20: 67. |
| 6. | Wang JG, Pan L, Wu B, Wang M. Familial characteristics of kidney-Yang deficiency and cold syndrome. J Toxicol Environ Health A 2006; 69: 1939-50. |
| 7. | Zhou LP, Liu WW, Zhang TE, et al. Resequencing DCDC5 in the flanking region of an LD-SNP derived from a Kidney-Yang deficiency syndrome family. Evid Based Complement Alternat Med 2011; 2011: 1-7. |
| 8. | Su M, Chen X, Chen Y, et al. UCP2 and UCP 3 variants and gene-environment interaction associated with prediabetes and T2DM in a rural population: a case control study in China. BMC Med Genet 2018; 19: 43. |
| 9. | Zhang T, Li J, Yu H, et al. Meta-analysis of GABRB 2 polymorphisms and the risk of schizophrenia combined with GWAS data of the Han Chinese population and psychiatric genomics consortium. PLoS One 2018; 13: e198690. |
| 10. | Wang X, Sun B, Jiang Q, et al. mRNA m(6)A plays opposite role in regulating UCP2 and PNPLA2 protein expression in adipocytes. Int J Obes (Lond) 2018; 42: 1912-24. |
| 11. | Hu XQ, Zhou Y, Chen J, et al. DNA Methylation and transcription of HLA-F and serum cytokines relate to Chinese medicine syndrome classification in patients with chronic hepatitis B. Chin J Integr Med 2022; 28: 501-8. |
| 12. | Hu ZR, Zheng WJ, Yan Q, Hu WW, Sun XS. Bioinformatic analysis of differentially expressed genes and Chinese medicine prediction for ulcerative colitis. Zhong Guo Zhong Yao Za Zhi 2020; 45: 1684-90. |
| 13. | Qiao C, Gu C, Wen S, et al. The integrated bioinformatic assay of genetic expression features and analyses of Traditional Chinese Medicine specific constitution reveal metabolic characteristics and targets in steatosis of nonalcoholic fatty liver disease. Hepat Med 2023; 15: 165-83. |
| 14. |
De Carvalho TR, Giaretta AA, Teixeira BF, Martins LB. New bioacoustic and distributional data on Bokermannohyla sapiranga Brandão et al., 2012 (Anura: Hylidae): revisiting its diagnosis in comparison with B. pseudopseudis (Miranda-Ribeiro, 1937). Zootaxa 2013; 3746: 383-92.
DOI PMID |
| 15. | Chen Y, Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res 2020; 48: D127-31. |
| 16. | Mon-Lopez D, Tejero-Gonzalez CM. Validity and reliability of the TargetScan ISSF Pistol & Rifle application for measuring shooting performance. Scand J Med Sci Sports 2019; 29: 1707-12. |
| 17. | Furió-Tarí P, Tarazona S, Gabaldón T, Enright AJ, Conesa A. spongeScan: a web for detecting microRNA binding elements in lncRNA sequences. Nucleic Acids Res 2016; 44: W176-80. |
| 18. |
Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015; 12: 453-7.
DOI PMID |
| 19. |
Mandrekar JN. Receiver operating characteristic curve in diagnostic test assessment. J Thorac Oncol 2010; 5: 1315-6.
DOI PMID |
| 20. |
Yu R, Liu D, Yang Y, et al. Expression profiling-based clustering of healthy subjects recapitulates classifications defined by clinical observation in Chinese medicine. J Genet Genomics 2017; 44: 191-7.
DOI PMID |
| 21. | Zhang C, Tam CW, Tang G, Chen Y, Wang N, Feng Y. Spatial transcriptomic analysis using R-based computational machine learning reveals the genetic profile of Yang or Yin deficiency syndrome in Chinese medicine theory. Evid Based Complement Alternat Med 2022; 2022: 5503181. |
| 22. | Gilca M, Stoian I, Gaman L. A new insight into estrogen signaling: Yin-Yang perspective. J Altern Complement Med 2013; 19: 63-8. |
| 23. | O'Connor GM, McVicar D. The Yin-Yang of KIR3DL1/S1: molecular mechanisms and cellular function. Crit Rev Immunol 2013; 33: 203-18. |
| 24. | Wang GZ, Goff SP. Regulation of Yin Yang 1 by tyrosine phosphorylation. J Biol Chem 2015; 290: 21890-900. |
| 25. | Zhang WJ, Wu XN, Shi TT, et al. Regulation of transcription factor Yin Yang 1 by SET7/9-mediated lysine methylation. Sci Rep 2016; 6: 21718. |
| 26. |
Rissman EF. Roles of oestrogen receptors alpha and beta in behavioural neuroendocrinology: beyond Yin/Yang. J Neuroendocrinol 2008; 20: 873-9.
DOI PMID |
| 27. | Huang C, Chen Y, Li B, et al. Traditional Chinese medicine constitution discrimination model based on metabolomics and random forest decision tree algorithm. Evid Based Complement Alternat Med 2022; 2022: 3490130. |
| 28. |
Liang X, Wang Q, Jiang Z, et al. Clinical research linking Traditional Chinese Medicine constitution types with diseases: a literature review of 1639 observational studies. J Tradit Chin Med 2020; 40: 690-702.
DOI |
| 29. | Nie X, Wang C, Zhang H, et al. The original scores of traditional Chinese medicine constitutions are risk and diagnostic factors in middle-aged and older adults with sarcopenia. Aging Med (Milton) 2024; 7: 334-40. |
| 30. | Nguyen MT, Min KH, Kim D, Park SY, Lee W. CFL 2 is an essential mediator for myogenic differentiation in C2C12 myoblasts. Biochem Biophys Res Commun 2020; 533: 710-6. |
| 31. | Zhang Z, Chuang Y, Ke X, et al. The influence of TCM constitutions and neurocognitive function in elderly Macau individuals. Chin Med 2021; 16: 32. |
| 32. | Zhu S, Xu N, Han Y, et al. MTERF 3 contributes to MPP+-induced mitochondrial dysfunction in SH-SY5Y cells. Acta Biochim Biophys Sin (Shanghai) 2022; 54: 1113-21. |
| 33. | Yu R, Liang J, Liu Q, Niu XZ, Lopez DH, Hou S. The relationship of CCL4, BCL2A1, and NFKBIA genes with premature aging in women of Yin deficiency constitution. Exp Gerontol 2021; 149: 111316. |
| 34. |
Braden K, Campolo M, Li Y, et al. Activation of GPR183 by 7α,25-dihydroxycholesterol induces behavioral hypersensitivity through mitogen-activated protein kinase and nuclear factor-κB. J Pharmacol Exp Ther 2022; 383: 172-81.
DOI PMID |
| 35. | Li YS, Li Y. Progress in the study of Yang-deficiency constitution in terms of Traditional Chinese Medicine: a narrative review. J Tradit Chin Med 2023; 43: 409-16. |
| 36. |
Jia D, Huang L, Bischoff J, Moses MA. The endogenous zinc finger transcription factor, ZNF24, modulates the angiogenic potential of human microvascular endothelial cells. FASEB J 2015; 29: 1371-82.
DOI PMID |
| 37. | Shuxian S, Xiuping Z, Jiayi M, et al. Serum protein profile of yang-deficiency constitution in Traditional Chinese Medicine revealed by protein microarray analyses. J Tradit Chin Med Sci 2019; 6: 67-74. |
| 38. | Fernandes HB, Oliveira BDS, Machado CA, et al. Behavioral, neurochemical and neuroimmune features of RasGEF1b deficient mice. Prog Neuropsychopharmacol Biol Psychiatry 2024; 129: 110908. |
| 39. | Bogucka-Janczi K, Harms G, Coissieux MM, Bentires-Alj M, Thiede B, Rajalingam K. ERK3/MAPK 6 dictates CDC42/RAC1 activity and ARP2/3-dependent actin polymerization. Elife 2023; 12: e85167. |
| 40. | Yang SH, Sharrocks AD, Whitmarsh AJ. MAP kinase signalling cascades and transcriptional regulation. Gene 2013; 513: 1-13. |
| 41. | Cai Q, Zhou W, Wang W, et al. MAPK6-AKT signaling promotes tumor growth and resistance to mTOR kinase blockade. Sci Adv 2021; 7: eabi6439. |
| 42. | Gil-Jaramillo N, Aristizábal-Pachón AF, Luque Aleman MA, et al. Competing endogenous RNAs in human astrocytes: crosstalk and interacting networks in response to lipotoxicity. Front Neurosci 2023; 17: 1195840. |
| 43. | Komai T, Inoue M, Okamura T, et al. Transforming growth Factor-β and interleukin-10 synergistically regulate humoral immunity via modulating metabolic signals. Front Immunol 2018; 9: 1364. |
| 44. | Tylutka A, Walas Ł, Zembron-Lacny A. Level of IL-6, TNF, and IL-1β and age-related diseases: a systematic review and meta-analysis. Front Immunol 2024; 15: 1330386. |
| 45. | Yan Q. The Yin-Yang dynamics in cancer pharmacogenomics and personalized medicine. Methods Mol Biol 2022; 2547: 141-63. |
| 46. | Zhu X. Seeing the Yin and Yang in cell biology. Mol Biol Cell 2010; 21: 3827-8. |
| 47. | Tang L, Chen L, Ye C, et al. Population-based associations between progression of normal-tension glaucoma and Yang-deficient constitution among Chinese persons. Br J Ophthalmol 2023; 107: 37-42. |
| 48. |
Chen Y, Wu Y, Yao H, et al. miRNA Expression profile of saliva in subjects of Yang deficiency constitution and Yin deficiency constitution. Cell Physiol Biochem 2018; 49: 2088-98.
DOI PMID |
| 49. | Yuan C, Zhang W, Wang J, et al. Chinese medicine phenomics (Chinmedphenomics): personalized, precise and promising. Phenomics 2022; 2: 383-8. |
| 50. |
Allam-Ndoul B, Guénard F, Barbier O, Vohl MC. Effect of n-3 fatty acids on the expression of inflammatory genes in THP-1 macrophages. Lipids Health Dis 2016; 15: 69.
DOI PMID |
| 51. | Madaniyazi L, Li S, Li S, Guo Y. Candidate gene expression in response to low-level air pollution. Environ Int 2020; 140: 105610. |
| 52. | von Voss L, Arora T, Assis J, et al. Sexual dimorphism in the immunometabolic role of Gpr 183 in mice. J Endocr Soc 2024; 8: bvae188. |
| 53. | Zhai Y, Li J, Cao Y, et al. Gut microbial composition associated with risk of premature aging in women with Yin-deficiency constitution. Front Cell Infect Microbiol 2025; 14: 1500959. |
| 54. | Huang D, Chen L, Ji Q, et al. Lead aggravates Alzheimer's disease pathology via mitochondrial copper accumulation regulated by COX17. Redox Biol 2024; 69: 102990. |
| 55. | Ma G, Li C, Ji P, et al. Association of Traditional Chinese Medicine body constitution and cold syndrome with leukocyte mitochondrial functions: an observational study. Medicine (Baltimore) 2023; 102: e32694. |
| [1] | LI Tianxing, ZHU Linghui, WANG Xueke, TANG Jun, YANG Lingling, PANG Guoming, LI Huang, WANG Liying, DONG Yang, ZHAO Shipeng, LI Yingshuai, LI Lingru. Gut microbial characteristics of the damp-heat constitution: a population-based multicenter cross-sectional study [J]. Journal of Traditional Chinese Medicine, 2025, 45(1): 140-151. |
| [2] | ZHOU Ying, LI Ping, LUAN Jianwei, SHEN Rui, WU Yinglan, XU Qiwen, WANG Xinyue, ZHU Yao, XU Xiangru, LIU Zitian, JIANG Yuning, ZHONG Yong, HE Yun, JIANG Weimin. Study on blood pressure rhythm in hypertensive patients with Yin deficiency syndrome and a random forest model for predicting hypertension with Yin deficiency syndrome [J]. Journal of Traditional Chinese Medicine, 2024, 44(3): 564-571. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||

